Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Short description of portfolio item number 1
Short description of portfolio item number 2 
Published in Phys. Rev. E, 2018
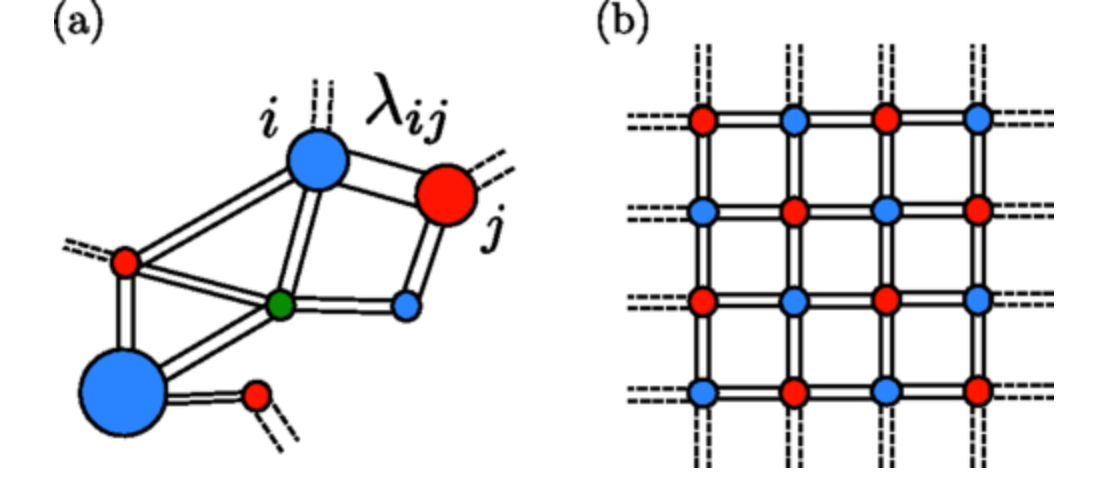
The exchange of diffusive metabolites is known to control the spatial patterns formed by microbial populations, but most laboratory conditions do not replicate the complex geometry of natural habitats. We mathematically model mutualistic interactions within a minimal unit of structure: two growing reservoirs linked by a diffusive channel through which metabolites are exchanged. Analytical and numerical solutions of the model, parametrized on a defined algal-bacterial mutualism, predict the necessary establishment conditions and how this depends, often counterintuitively, on diffusion geometry. We connect our findings to understanding complex behavior in synthetic and naturally occurring microbial communities.
Download here
Published in New Phytologist, 2018
Many algae, such as the Chlamydomonas relative, L. rostrata, require vitamin B12 for growth, which can be provided by certain bacteria in their environment. We used iTRAQ proteomics to determine the L. rostrata proteome when grown axenically (with B12 supplementation) or in coculture with the bacterium Mesorhizobium loti. Amino acid biosynthesis was higher, but photosynthesis proteins and electron transfer lower in coculture indicating that despite the stability of the mutualism, L. rostrata experiences stress in coculture and may adjust its metabolism accordingly.
Download here
Published in New Phytologist, 2019
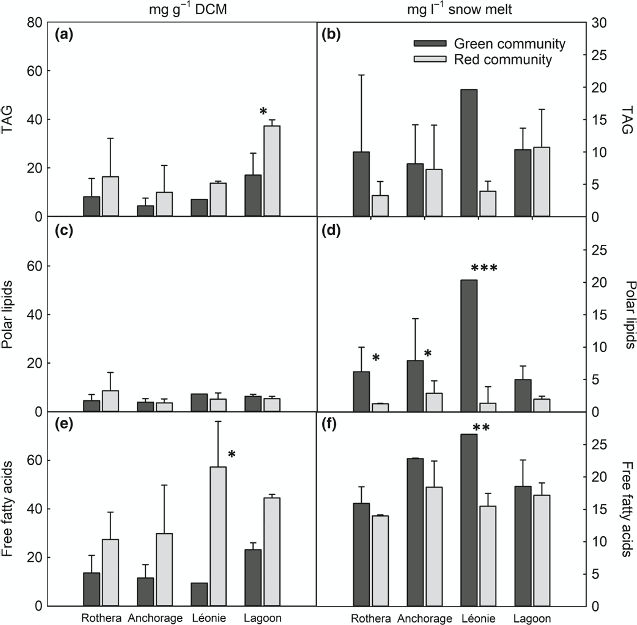
Snow algae are found in snowfields across cold regions of the planet, forming highly visible red and green patches below and on the snow surface. In Antarctica, they contribute significantly to terrestrial net primary productivity due to the paucity of land plants, but our knowledge of these communities is limited. Here we provide the first description of the metabolic and species diversity of green and red snow algae communities from four locations in Ryder Bay (Adelaide Island, 68°S), Antarctic Peninsula.
Download here
Published in PhD diss., University of Cambridge, 2019., 2019

My PhD thesis: Vitamin B12 is synthesised only by prokaryotes yet is widely required by eukaryotes as an enzyme cofactor. Roughly half of all algae require vitamin B12, and the phylogenetic distribution of this trait suggests that it has evolved on multiple occasions. Previous work using artificial evolution generated a metE mutant of Chlamydomonas reinhardtii (hereafter metE7) that requires B12 for growth. Here, I use metE7 to investigate how a newly-evolved B12 auxotroph might cope with B12 limitation and interact with B12-producing bacteria. This work has provided an insight into the challenges of evolving B12 auxotrophy making it all the more remarkable that it is such a common trait.
Download here
Published in Proceedings of the Machine Learning for Health NeurIPS Workshop, 2020
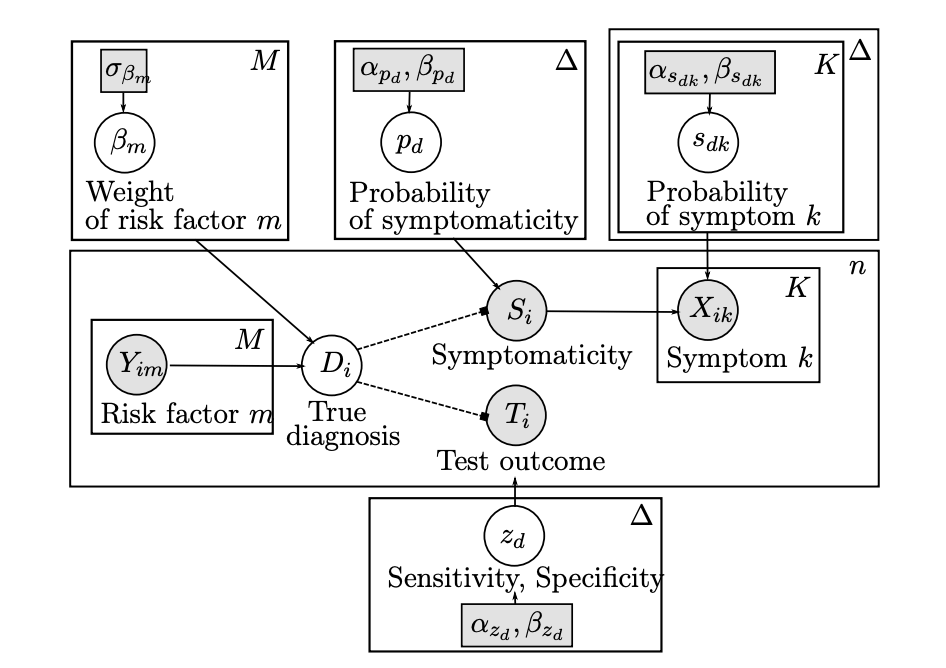
Medical practitioners often reach a diagnosis after combining a patient’s medical history, their answers to certain questions, and the results of medical tests. We sought to apply this framework in an automated manner using online questionnaires and lateral flow test results. We devised a Stochastic Expectation-Maximization algorithm within a Bayesian formalism to combine heterogeneous data sources, quantify their uncertainty and produce a probabilistic diagnosis. We quantify the potential of this approach on simulated data, and showcase its practicality by deploying it on a real COVID-19 immunity study.
Download here
Published in Plant physiology, 2020
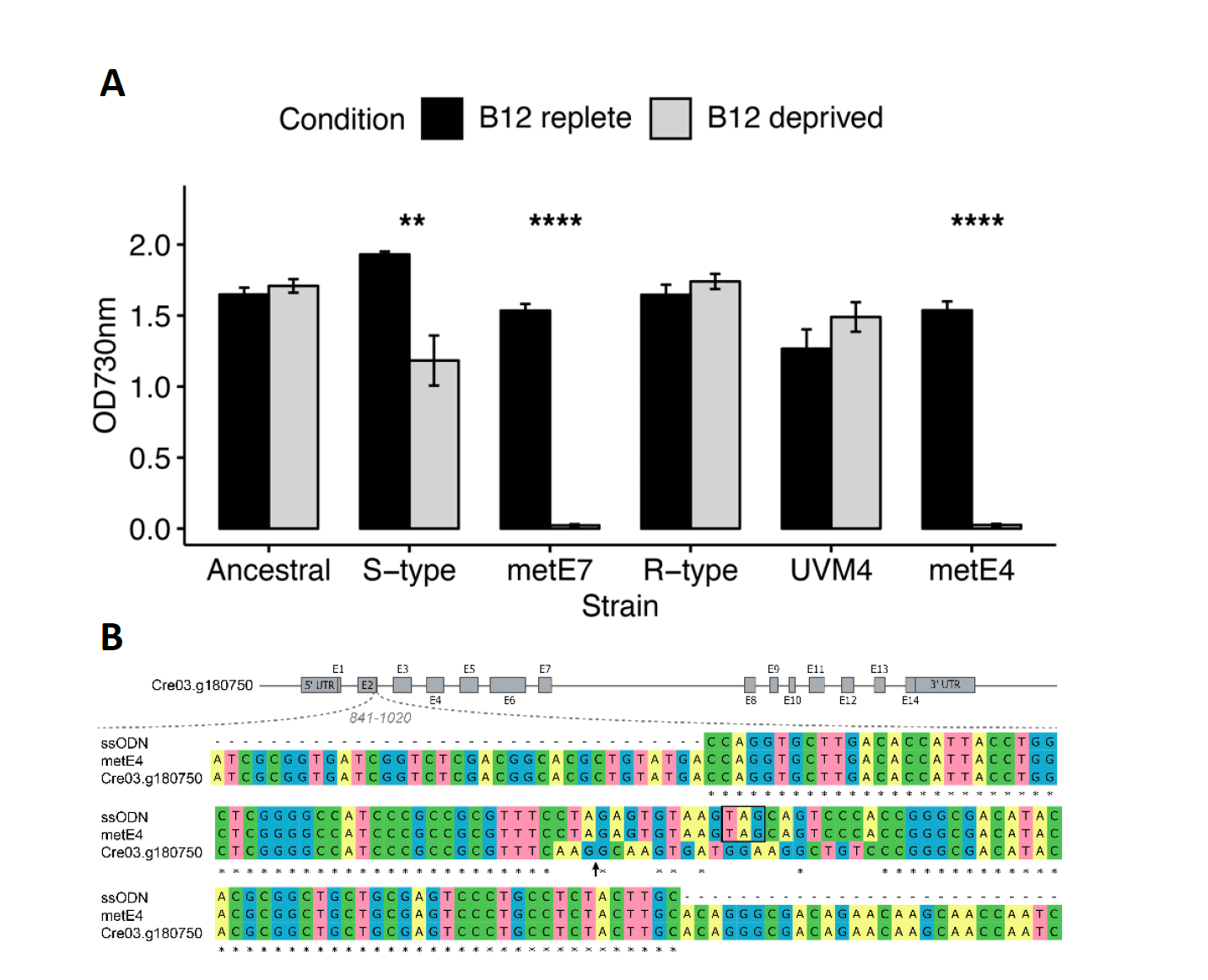
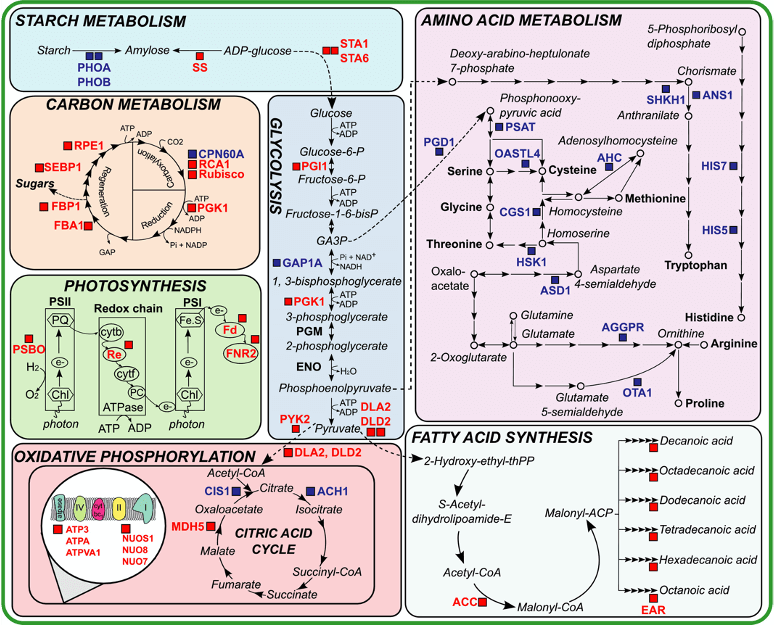
Microalgae have evolved B12 dependence on multiple occasions, and we previously demonstrated that Chlamydomonas reinhardtii can evolve B12 dependence in the lab. This clone provides a unique opportunity to study the physiology of a nascent B12 auxotroph. Our analyses demonstrate that B12 deprivation of this strain disrupts C1 metabolism, causes an accumulation of starch and triacylglycerides, and leads to a decrease in photosynthetic pigments, proteins, and free amino acids. We exposed this strain to further experimental evolution with reduced B12 levels and in coculture with bacteria and showed these lines were better able to survive B12 limitation.
Download here
Published in PloS One, 2021
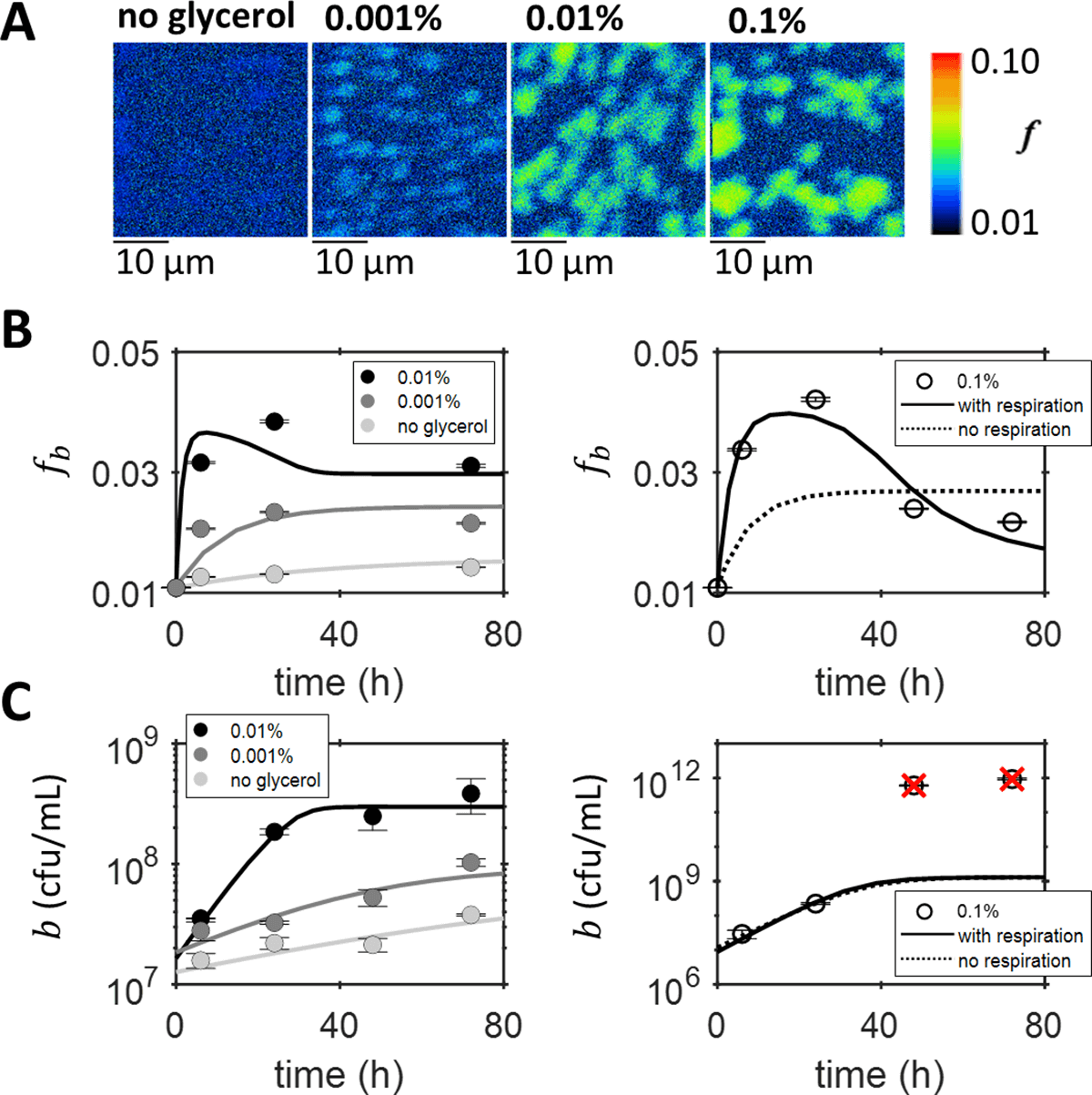
A key feature of microbial interactions is the exchange of nutrients between cells. Here we present a novel approach that combines isotope labelling and secondary isotope mass spectrometry experiments with mechanistic modelling to reveal otherwise inaccessible nutrient kinetics. Studying the establishment of a symbiosis between a B12-dependent alga and B12-producing bacteria, our results suggest that the onset of mutualistic cross-feeding is delayed by pre-existing nutrient pools. Our method is widely applicable to other microbial systems, and will contribute to furthering a mechanistic understanding of microbial interactions.
Download here
Published in JMIR public health and surveillance, 2021
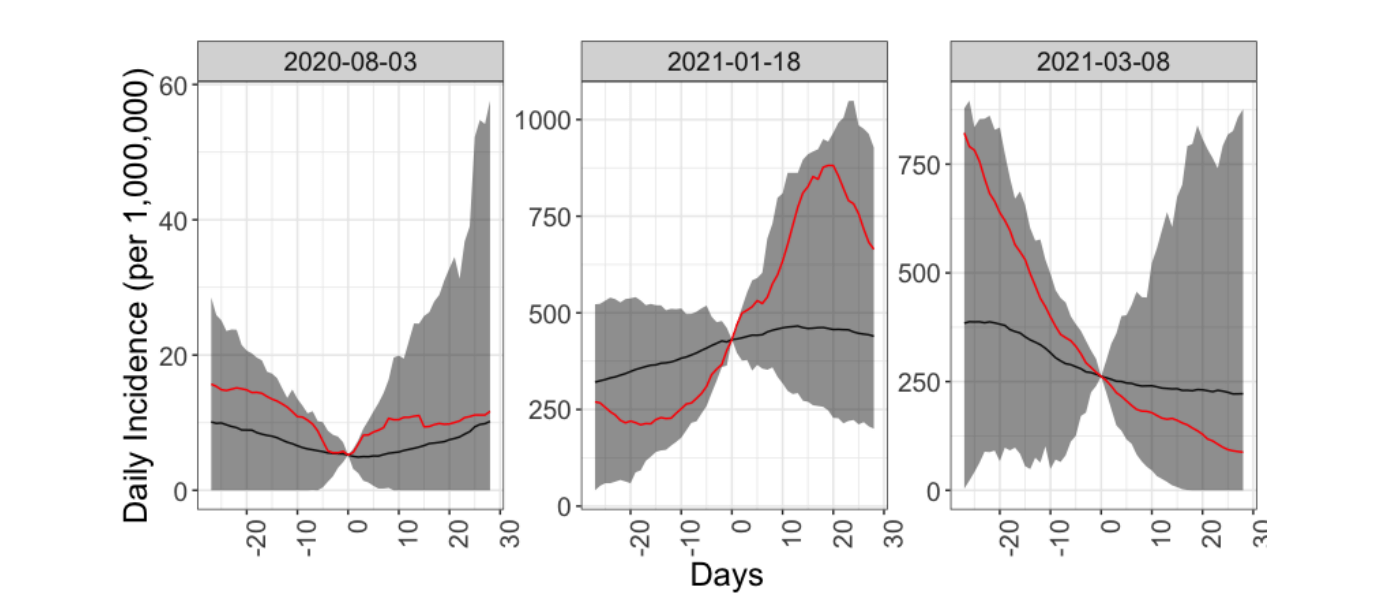
Predicting infectious disease transmission at future public events is a highly uncertain process. In the context of the COVID-19 pandemic, we aimed to systematize the process by 1) reliably model the infectious proportion of participants at a future event, 2) evaluate the efficiency of pre-event screening protocols, and 3) model an event’s transmission dynamics and uncertainty using Monte Carlo simulations. Illustrating the process on a hypothetical concert at the Royal Albert Hall, we tested the effect of factors such as mask wearing, participant reduction, antigen test screening, and delaying the event. Our model is available as a user-friendly RShiny interface and can give event organizers and participants alike an rough understanding of the infection risks associated with attending public events.
Download here
Published in Applied and Environmental Microbiology, 2022
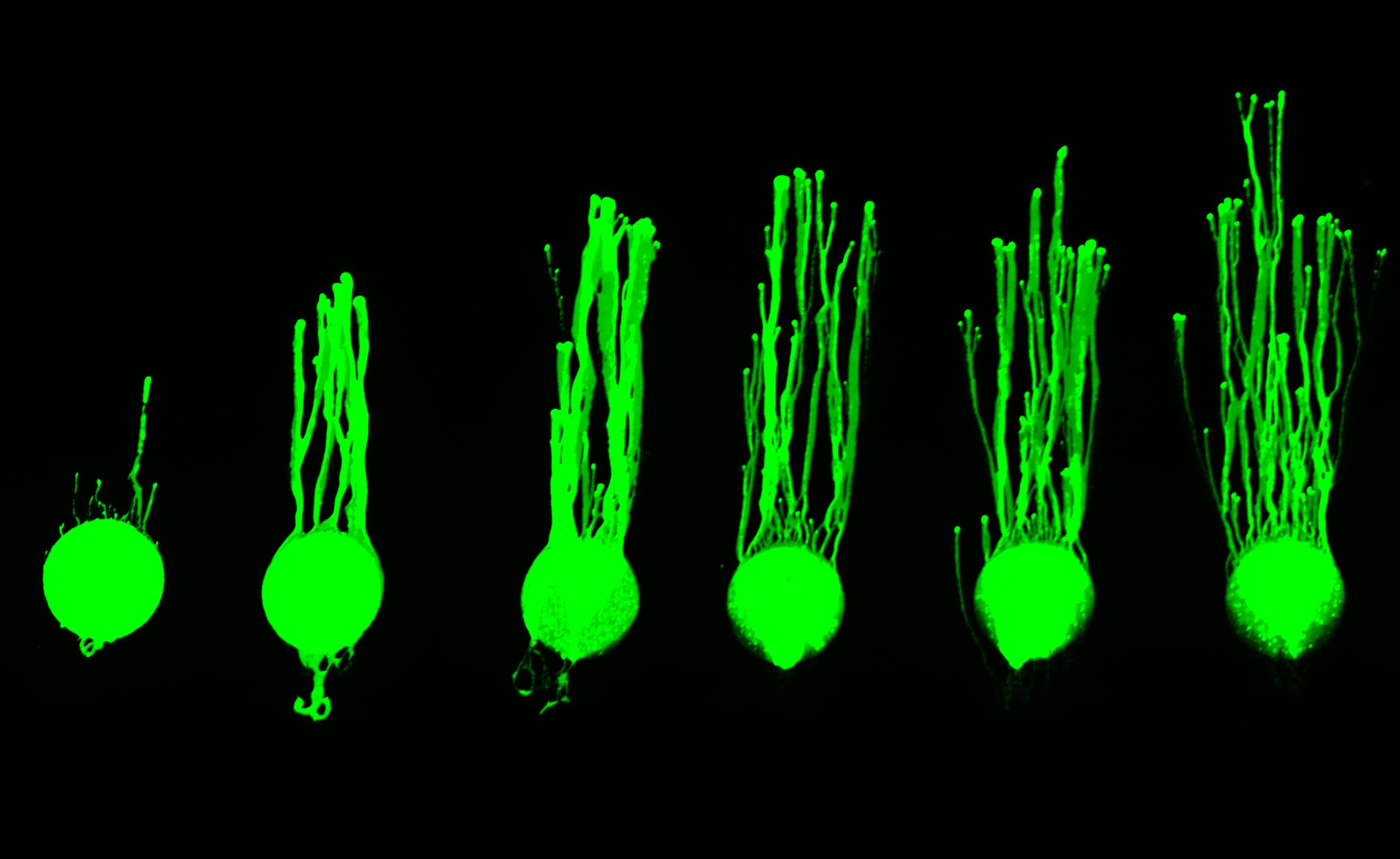
Some photosynthetic microbes have evolved a sophisticated process called phototaxis to move toward or away from a light source and so optimize their light exposure. In many hot springs in Yellowstone National Park, cyanobacteria thrive in thick, laminated biofilms or microbial mats, where small movements can result in large changes in light exposure. We found that the two most abundant cyanobacterial ecotypes in these springs exhibited unexpected differences in their speed, directionality, and responses to different intensities or qualities of light. Our results reveal that diverse phototactic strategies can exist even among close relatives in the same environment.
Download here
Published:
Practical class, Cambridge, Department of Plant Sciences, 2016
I helped to organise and run undergraduate practicals for two courses in the Natural Sciences Tripos. These involved helping students design experiments, perform PCR reactions, assay enzyme activity and use microscopy to investigate plant traits.
Practical class, Cambridge, Department of Plant Sciences, 2016
I designed a research project with Bradford Loh to investigate the phenotype of Chlamydomonas nivalis, a species of Antartic snow algae. Bradford really enjoyed the project and did great work which ended up being published in the New Phytologist.
Undergraduate research student, Cambridge, Department of Plant Sciences, 2017
I was recommended to tutor two cohorts of Trinity college undergraduates studying the Physiology of Organisms course in the Natural Sciences Tripos. This was a fantastic experience of discussing and explaining Plant Physiology concepts to very capable and diligent students.
Practical class, Cambridge, Department of Plant Sciences, 2017
I designed a research project with Sam Fitzsimmons to study interactions and signalling between B12-dependent algae and bacteria. Sam was a great person to have as a mentee, always very positive and inquisitive, and his project helped shape some of our labs research on B12-based mutualisms.
Undergraduate research student, Stanford, 2022
The BioBUDS course was a new course at Stanford for Building Up Developing Scientists, particularly for people with minimal research experience and from underrepresented backgrounds. Ronan Esperanza was a great mentee who worked hard and displayed a good deal of scientific creativity during the 10-week program. We worked together on understanding more about the way in which thermophilic cyanobacteria respond dynamically to changing light intensity and quality. Ronan produced a fantastic presentation on his work which drew the attention of many of his peers.
Summer student, Carnegie Institution for Science, Department of Plant Biology, 2022
Dr. Victoria Calatrava and I designed a research project with Alicia Sanoyca, an undergraduate from UCLA. Alicia will be investigating physical interactions between Synechococcus and Chloroflexus as well as the motility of these species. We hope that Alicia will enjoy the experience and learn a lot about microbiological research during the summer internship.